Identical by descent
From ISOGG Wiki
(Redirected from Identical By Descent segment)Identical by descent (IBD) is a term used in genetic genealogy to describe a matching segment of DNA shared by two or more people that has been inherited from a common ancestor without any intervening recombination. The segments are considered to match if all the alleles on a paternal or maternal chromosome are identical (barring rare mutations and genotyping errors) and if the minimum threshold conditions set by the testing company have been met.
Everyone has two copies of each chromosome – one chromosome inherited from their father and one chromosome inherited from their mother. Matching segments can be on half-identical regions (HIRs) (matches on the paternal or maternal chromosome) or fully identical regions (FIRs) (matches on both the paternal and maternal chromosome). FIRs are generally only seen in full siblings and double cousins, but are sometimes found in more distant relatives if the individual comes from an endogamous (intermarrying) population.
The length of an IBD segment can be measured in centiMorgans (a unit of genetic distance) or in megabases (a unit of physical distance). The three major autosomal DNA testing companies (23andMe, AncestryDNA and Family Tree DNA) all now report segment sizes in centiMorgans. AncestryDNA originally used megabases for their matching algorithms but converted to centiMorgans in about January 2014. Both 23andMe and Family Tree DNA provide information on the matching segments which can be downloaded into a spreadsheet, and they also provide chromosome browsers which allow customers to see a visual representation of the matching half-identical DNA segments. The chromosome browsers show only one chromosome from each pair of chromosomes and are unable to distinguish between the maternally inherited and paternally inherited chromosomes. AncestryDNA do not currently provide a chromosome browser and do not provide access to the underlying matching segment data.
The segment boundaries are somewhat fuzzy. 23andMe round the segment length to the nearest tenth of a centiMorgan and round the segment start and end co-ordinates to the closest millionth base pair to reflect the uncertainty in the exact locations of the segment boundaries. Family Tree DNA round the start and stop positions to the nearest complete block of 100 SNPs on each end of the matching segment.
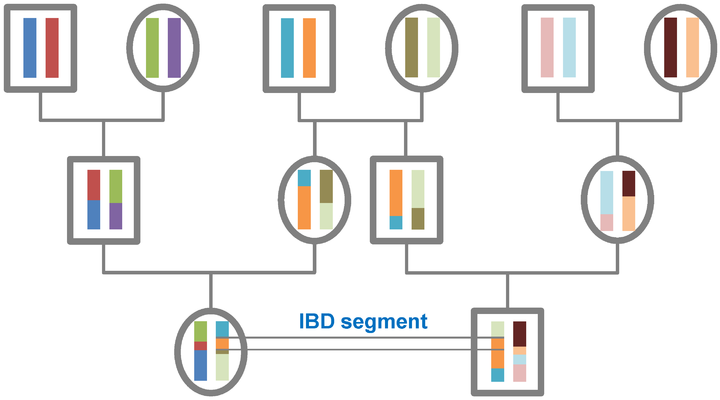
Identity by descent can be considered on various timescales. According to population genetics theory all individuals have common ancestry in the distant past, and we all have short, old IBD segments in common. For the purposes of genetic genealogy the focus is on detecting large IBD segments within a genealogical timeframe (effectively within the last ten generations) where there is a possibility of identifying the common ancestor through documentary records. In general terms the larger the segment the closer the relationship, but the frequency of the segment also needs to be taken into account. High-frequency IBD segments are more likely to be a signal of distant sharing at the population level whereas a segment that is only observed in two independently sampled individuals is more likely to be IBD.[1]
Any given pair of individuals is related through many common ancestors, though many of these relationships will be too distant to result in detectable IBD segments. If the two individuals have ancestors from the same geographical region they might have many recent common ancestors, but many of the relationships will not result in IBD sharing, and there might only be one or two segments inherited from just a few of their many common ancestors. In a study of a European subset of the Population Reference Sample (POPRES) dataset it was estimated that for the most part IBD blocks longer than 4 cM come from 500 to 1,500 years ago, and blocks longer than 10 cM are within the last 500 years.[2]
In general it will be difficult to find the genealogical connection with the majority of your matches under about 15 cMs. Most people are unable to trace all of their ancestral lines back ten generations or so, and the common ancestral couple cannot therefore be identified. Even if a shared ancestral couple can be identified, without tracing all the other ancestral lines you cannot eliminate the possibility of shared ancestry on other as yet undocumented lines. Many matches under 15 cMs will in any case share ancestry more than ten generations ago and will be mostly beyond the reach of genealogical records.[3]
A study by Speed and Balding (2014) using computer simulations going back for 50 generations showed that over 50% of 5 mB segments date back over 20 generations, and fewer than 40% of 10 mB segments are within the last 10 generations. Larger segments can still date back quite some time and it was found that around 40% of 20 mB segments date back beyond 10 generations.[4] Figure 2 from this paper showing the distribution of IBD segments of differing sizes by age is reproduced below. (Note that these simulations were based on megabases. As a general rule of thumb one megabase is equivalent to one centiMorgan.)

False positive matches
The companies' matching algorithms do not treat the paternal and maternal chromosomes separately. Consequently consecutive SNP results for a short segment of DNA may appear to be half-identical in two individuals when in actuality the DNA sequences are not identical because the SNPs match on opposing chromosomes or because of errors in the matching algorithms. False matches can be the result of pseudosegments (matching alleles zig-zagging backwards and forwards between the maternal side and the paternal side, also called haplotype switching), compound segments and fuzzy boundaries.[5][6] For a good illustration and explanation of a pseudosegment (also known as a spurious segment, an erroneous segment or a phantom segment) see Don Worth's diagram.
A simple check on the false positive/negative match rate can be made by looking at the percentage of matches received by a child which do not appear in the match list of either parent. False positive matching rates of between 12% and 23% have been reported for Family Finder data before their move to an Illumina GSA microarray, and up to 34% at Ancestry with the algorithm in use prior to increasing the segment minimum to 8 cM.[7][8][9] However, the lack of matches in some cases might be the result of false negatives. Nevertheless, it is important to remember that if only one parent has been tested, one should not automatically assume that all the remaining matches can be assigned to the other parent. A significant proportion will be non-IBD.
False positive matches are more likely to be seen in unphased data (phasing is the process of assigning alleles to the mother or the father.) The highest degree of accuracy is achieved by using the phased data from a two-parent/one child trio, where the error rate for phasing is only 0.01%. In the absence of trio data it is possible to phase data by inference using samples from reference populations. This is known as statistical phasing, computational phasing or algorithm-based phasing. AncestryDNA is currently the only company to phase all the customer data prior to matching. Ancestry uses a proprietary phasing algorithm known as Underdog. When phased results were compared with a test set of 1,188 trio-phased samples Underdog had a phase error of 0.93% and an impute error of 1.09%.[10] In theory Ancestry should have a lower false matching rate than 23andMe and FTDNA, though there are still likely to be false positives, particularly for short phased segments in the 5-8 cM range. None of the the companies currently offer the facility for customers who have tested both parents to phase their own trio data. However, trio phasing can be done by uploading results to the free third-party website GedMatch.
In general, the larger the shared segments the more likely that the match is valid. Valid segments under 7 cM cannot be reliably detected with the currently available genetic genealogy tests. Durand et al (2014) analysed phased data from 2,952 father-mother-child trios in the 23andMe dataset and identified a false positive rate of over 67% for 2–4 cM segments. They reported that "most 2–3 cM segments are erroneous and only segments longer than 5 cM have a negligible number of false positives".[11] The error rate is likely to be much higher in unphased data. One Family Tree DNA customer manually phased data from 14 family members and found that 85% of the segments under 5 cMs were false matches.[12] This figure is consistent with the findings of John Walden (see table below) who found that 86% of half identical regions under 5 cMs were false matches when both sides were phased. A similar rate was reported by Tim Janzen (see second table below). He found that 79.7% of the half-identical matches under 5 cMs for both he and his wife were false matches with only one side phased. As the false positive rate for segments under 5 cMs is so high these segments should generally not be used in genealogical analyses.
The following table shows the percentage of matching segments (half-identical regions) that survived phasing. The results are based on an analysis of 9000 half-identical regions by John Walden as reported to Tim Janzen.[13][14]
| cM | % IBD one side phased | % IBD both sides phased | % lost by second phasing |
|---|---|---|---|
| 15 | 100 | 100 | 0 |
| 13 | 98 | 99 | 1 |
| 12 | 98 | 97 | 1 |
| 11 | 94 | 90 | 4 |
| 10 | 89 | 86 | 4 |
| 9 | 85 | 80 | 6 |
| 8 | 78 | 62 | 23 |
| 7 | 63 | 42 | 33 |
| 6 | 42 | 26 | 38 |
| 5 | 29 | 14 | 52 |
| 4 | 18 | 5 | 72 |
| 3 | 9 | 1 | 89 |
Note that misreadings of this table have not been uncommon. The center column titled "% IBD both sides phased" indicates the percentage of the time segments of a particular size were still accounted for after traditional trio phasing. In other words, 62% of 8 cM segments could be accounted for; 42% of 7 cM segments could be accounted for; 26% of 6 cM segments could be accounted for, and so on. The other 38%, 58%, and 74%, respectively, of the reported segments from the child's test results were not present in the results from either parent and thus could be deemed as false positive.
A logistical regression equation was developed from the above data by A.J. Levin using the SDSFoundations plugin for R. One should be able to use this equation to estimate the probability of an unphased match disappearing or remaining after phasing, for any length of match.
The following analysis is provided by Tim Janzen. The table shows a comparison of Tim Janzen’s and Rachel Janzen’s matches at GEDmatch at various thresholds using the phased versus unphased Family Finder data.[13]
| 20 cM | 15 cM | 11 cM | 10 cM | 9 cM | 8 cM | 7 cM | 6 cM | 5 cM | |
|---|---|---|---|---|---|---|---|---|---|
| TJ unphased | 21 | 54 | 183 | 245 | 421 | 870 | 1813 | 3512 | 8716 |
| TJ phased | 21 | 54 | 180 | 175 | 263 | 351 | 542 | 1015 | 1713 |
| RJ unphased | 10 | 27 | 98 | 149 | 349 | 1042 | 2331 | 3673 | 7460 |
| RJ phased | 10 | 27 | 95 | 98 | 145 | 197 | 326 | 832 | 1564 |
| % IBD | 100.0 | 100.0 | 97.9 | 69.3 | 53.0 | 28.7 | 20.9 | 25.7 | 20.3 |
Thresholds for matches
Each company sets their own autosomal DNA match thresholds. These criteria must be met before the company will report that two individuals very likely inherited their half-identical matching segments from a recent common ancestor.
Excess IBD sharing
There are some regions of the genome that are prone to excess IBD sharing. These regions are known colloquially as pile-up regions.
The human leukocyte antigen (HLA) region on chromosome 6 has been found to exhibit a much higher amount of IBD sharing than expected both within and across populations. The genes in this region are responsible for regulation of the immune system in humans, and it is thought that favourable mutations have developed as a result of natural selection.[15] Gusev et al (2011) reported that in Ashkenazi Jews “the entire segment of chromosome 6, between 25 and 35 Mb, is shared among individuals unrecombined at least 4-fold more than any other region in the genome”. They also found that the HLA region had a very high sharing density in some of the other populations studied (Western European, Gujarati Indians, Luhya Kenyans and Yoruba Nigerians). [16] An excess of IBD sharing in the HLA region was also found in a study of the French founder population in Quebec.[17] For a good explanation of the underlying reasons for the increased IBD sharing in the HLA region of chromosome 6 see the article by Ann Turner (2010) Up hill and down dale in the genomic landscape: the odd distribution of matching segments in the Journal of Genetic Genealogy.[18]
Albrechtson et al (2011) found a region on chromosome 8 centred on position 9.9 Mb which exhibited an excess of IBD sharing. High IBD sharing signals were also seen on each side of the centromere on chromosome 11. These regions contain clusters of olfactory receptors. Evidence for increased IBD was also seen on chromosomes 2, 3, and 5.[15]
Gusev et al (2011) found recurrently shared regions in chromosomes 2, 4, and 8 in unrelated individuals of different geographical origins, and even across different continents of origin.[16] The region on chromosome 8 which was enriched for IBD sharing was in the location between 11.1 and 13.3 Mbs and in a different position to that reported by Albrechtson.
Li et al (2014) studied genetic relationships from whole genome sequencing data and identified a number of regions with excess IBD in both their pedigree and control dataset using three established methods for IBD detection (GERMLINE, fastIBD, and ISCA). The authors found that "the regions of spurious IBD were largely consistent between the three IBD methods and among European, East Asian, and Mexican American population which is a strong indication that the IBD segments in these regions are artifactual."[19] The excess IBD regions detected in the control populations are shown in the figure below.

The start and stop positions for the excess IBD regions identified by Li et al are shown in the table below.

Carmi et al (2014) analysed the whole genomes of 128 Ashkenazi Jews (AJs) and compared the samples with controls from a Flemish population. They found that there was a recent bottleneck in the Ashkenazi Jewish population of around 350 individuals 600–800 years ago. The authors phased the data for nearly all the chromosomes before performing their analysis. When they used a minimum segment length of 3 cMs they found that all AJs shared on average 1.85% of their genome. When they went up to 5 cMs they found that AJs shared on average 0.84% of their genome. Peaks of IBD sharing over 5 cMs in size were seen on chromosomes 9 and 19 in Ashkenazi Jews. However, the authors were unable to detect any "enrichment of sharing in the HLA region (≈chr 6:25-35 mB) as previously observed in Jewish and non-Jewish populations".[20]
AncestryDNA analysed the matches of over half a million US customers genotyped on an Illumina OmniExpress chip (~700,000 SNPs) and found that in certain regions of the genome hundreds and sometimes thousands of people were predicted to share DNA with each other. The problem was particularly pronounced in customers of Jewish ancestry and in some people of Hispanic descent, but the problem was observed across all ethnic groups. They concluded that these regions of identical DNA were likely to be attributed to a shared ethnic history rather than recent common ancestry.[21][22] Ancestry introduced new phasing and matching algorithms in November 2014 to remove the false matches resulting from these "pile-ups".[23] The technical details can be found in their DNA Matching White Paper “Discovering genetic matches across a massive, expanding genetic database”. However, the locations of the identified "pile-up" regions have not been disclosed.
See also the post Chromosome pile ups in genetic genealogy: examples from 23andMe and FTDNA on the Genealogy and Genomics blog.
Whole-genome sequencing
The terms IBD and IBS are more relevant to the results of SNP microarray testing than to results of whole-genome sequencing, because microarray testing provides so much less information per centiMorgan of DNA. Microarray test results have an additional complexity since they report on both copies of the chromosome, but the results (with the exception of AncestryDNA and MyHeritage) are not phased (that is, it is unknown which nucleotide is on which copy of the chromosome). Thus if one person's SNP result is (CC), this could be at least "half-identical" to either (CC) or (CT) in a second person. A homozygous mismatch such as (CC) vs. (TT) would be required before one could say the results are *not* identical.
Whole-genome sequencing (WGS) using next generation sequencing (NGS) technology, is not currently affordable for the genetic genealogy market, but is being used in academic studies: IBD tools are able to detect all 1st through 6th degree relationships and 55% of 9th through 11th degree relationships, a 5% to 15% increase in relationship detection compared to high-density microarray data.[19]
Whole-genome sequencing can also be used to detect more distant relationships for persons with 8th and 9th degrees of relationship by counting the total number of shared very rare SNPs.[24]
Further reading
- FTDNA defintion of identical by descent Family Tree DNA Learning Center
- Wheaton K. atDNA matches. Lesson 9 in the series "Beginners' Guide to Genetic Genealogy", Wheaton Surname Resources website, 2013.
- Turner A. "Satiable Curiosity: Identity Crisis: Identical by State or Identical by Descent?" Journal of Genetic Genealogy Fall 2011, Volume 7.
- Gregg Bonner's Family Finder pages Includes a link to a graphic of all his Family Finder matches on all chromosomes on both paternal and maternal side - arranged in filmstrip format
- Wikipedia article on identity by descent
Recommended blog posts
- Bettinger D. An in-depth analysis of the use of small segments as genealogical evidence. The Genetic Genealogist, 7 August 2022.
- Kennett D. Small segments and pile-ups. Cruwys News, 22 January 2018.
- Bettinger B. A small segment round-up. The Genetic Genealogist, 29 December 2017.
- Bettinger B. The effect of phasing on reducing false distant matches (or, phasing a parent using GEDmatch). The Genetic Genealogist 26 July 2017.
- Bettinger B. The danger of distant matches. The Genetic Genealogist, 6 January 2017. See also the additional analyses by Kevin Ireland and Ann Raymont.
- Turner A. Anatomy of an IBS segment. A guest post on Jim Bartlett's Segmentology blog, 2 October 2015.
- Granka J. Filtering DNA matches at AncestryDNA with Timber. Ancestry Blog, 8 June 2015.
- Kennett D. Tracking DNA segments through time and space. Cruwys News blog, 26 April 2015.
- Edwards D. Genealogy and autosomal DNA matches: common errors in proving an ancestor and the allure of easy gateway ancestors Genealogy and Genomics blog, 19 April 2015.
- Turner A. What a difference a phase makes. Guest post on The Genetic Genealogist Blog, 30 March 2015.
- Edwards D. Chromosome pile ups in genetic genealogy: examples from 23andMe and FTDNA. Genealogy and Genomics blog, 31 January 2015.
- Johnston K. What can the X chromosome tell us about the importance of small segments? Guest post on Kitty Cooper's blog, 19 December 2014.
- Bettinger B. Small matching segments - examining hypotheses. The Genetic Genealogist, 8 December 2014.
- Moore C. The folly of using small segments as proof in genealogical research. Part 1. Your Genetic Genealogist, 3 December 2014.
- Bettinger B. Small matching segments - friend or foe?. The Genetic Genealogist, 2 December 2014.
- Bettinger B. Goodbye false positives! AncestryDNA updates its matching algorithm. The Genetic Genealogist blog, 19 November 2014.
- Cooper K. When is a DNA segment match a real match? IBD or IBS or IBC?. Kitty Cooper's DNA Genealogy blog, 27 October 2014.
- Bettinger B. Finding genetic cousins - separating fact from fiction. The Genetic Genealogist, 15 October 2014. A preview of the methodology to be used to reduce the number of false positive matches at AncestryDNA.
- Coop G. How many genomic blocks do you share with a cousin?. Coop Lab blog, 2 December 2013.
- Coop G and Ralph P. Identification of genomic regions shared between distant relatives. The Coop Lab blog, 10 May 2013.
- Rose K. The ABCs of DNA - IBD vs IBS vs mIBC. DNA Genealogy blog, 30 January 2012. [Article snapshot from 2 May 2015.]
- Mount S. Genetic genealogy and the single segment. On Genetics blog 19 February 2011.
- Granka J. The DNA research and matching development life cycle. An explanation of the AncestryDNA matching process and their definitions of IBD and IBS.
Scientific papers
- Ying Q, Sannerud JG, Basu-Roy S, Hayward C, Williams AL (2021). Distinguishing Pedigree Relationships via Multi-Way Identity by Descent Sharing and Sex-Specific Genetic Maps. The American Journal of Human Genetics January; 108(1): 68-83.
- Zhou Y, Browning BL, Browning SR (2020). Population-Specific Recombination Maps from Segments of Identity by Descent. The American Journal of Human Genetics July; 107(1): 137–148.
- Seidman D., Shenoy SA, Kim M, et al (2020). Rapid, Phase-Free Detection of Long Identity-by-Descent Segments Enables Effective Relationship Classification. The American Journal of Human Genetics April; 106(4): 453–466.
- Ge J, Budowle B (2020). How Many Familial Relationship Testing Results Could Be Wrong? PLoS Genetics August; 16(8): e1008929.
- Browning SR, Browning BL (2020). Probabilistic Estimation of Identity by Descent Segment Endpoints and Detection of Recent Selection (preprint) BioRxiv, published online 15 July 2020: 2020.07.15.205179.
- Ramstetter MD, Dyer TD, Lehman DM et al (2017). Benchmarking relatedness inference methods with genome-wide data from thousands of relatives. Genetics published online on 24 July 2017.
- Bjelland DW, Lingala U, Patel PS, Jones M, Keller MC (2017). A fast and accurate method for detection of IBD shared haplotypes in genome-wide SNP data. European Journal of Human Genetics May; 25(5): 617-624. Epub 2017 Feb 8.
- Ramstetter M, Dyer TD, Lehman DM et al (2017). A performance assessment of relatedness inference methods using genome-wide data from thousands of relatives (preprint). BioRxiv, published online 4 February 2017.
- Povysil G, Hochreiter S (2016). IBD Sharing between Africans, Neandertals, and Denisovans. Genome Biology and Evolution 8 (12): 3406-3416.
- Fu, W, Browning SR, Browning BL, Akey JM (2016). Robust inference of identity by descent from exome-sequencing data. American Journal of Human Genetics 99 (5): 1106–1116.
- Chiang CWK, Ralph P, Novembre J (2016). Conflation of short identity-by-descent segments bias their inferred length distribution. G3 Genes Genomes Genetics 6: 1287.
- Fedorova L, Qiu S, Dutta R, Fedorov A (2016) Atlas of cryptic genetic relatedness among 1000 human genomes. Genome Biology and Evolution 8(3): 777-790.
- Rodriguez JM, Bercovici S, Huang L et al (2015). Parente2: a fast and accurate method for detecting identity by descent. Genome Research 25(2):280-9.
- Speed D, Balding DJ (2014). Relatedness in the post-genomic era. Nature Reviews Genetics. 16: 33-34. (subscription required). Authors' preprint available here
- Carmi S , Wilton PR , Wakeley J , Pe'er I (2014). A renewal theory approach to IBD sharing. Theoretical Population Biology 97: 35-48.
- Carmi S, Hui KY, Kochav E et al (2014). Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nature Communications 2014; 5: 4835. See in particular section 4.2 in the supplementary data pdf file where the authors discuss the IBD analysis.
- Durand EY, Eriksson N, McLean CY (2014). Reducing pervasive false positive identical-by-descent segments detected by large-scale pedigree analysis. Molecular Biology and Evolution 2014 doi: 10.1093/molbev/msu151. First published online: April 30, 2014. For a layman's summary see the 23andMe blog post:
- 23andMe scientists improve methods for finding relationships 23andMe blog, 12 May 2014.
- Li H, Glusman G, Hu H (2014). Relationship estimation from whole-genome sequence data. PLoS Genetics 2014 Jan 30;10(1):e1004144. Figure 1 indicates regions which are prone to excess IBD inference from WGS data.
- Hochreiter S (2013). HapFABIA: Identification of very short segments of identity by descent characterized by rare variants in large sequencing data. Nucleic Acids Research 2013 doi: 10.1093/nar/gkt1013.
- Carmi S, Palamara PF, Vacic V et al (2013). The variance of identity-by-descent sharing in the Wright–Fisher model. Genetics 193(3): 911-928.
- Su SY, Kasberger J, Baranzini S et al (2012). Detection of identity by descent using next-generation whole genome sequencing data. BMC Bioinformatics 13:121.
- Browning SR, Browning BL (2012). Identity by descent between distant relatives: detection and applications. Annual Review of Genetics 46: 617-33. A useful review article defining the terminology and summarising the different methodologies used to infer IBD segments.
- Thompson EA (2013). Identity by descent: variation in meiosis, across genomes, and in populations. Genetics 194(2): 301-326.
- Henn BM, Hon L, Macpherson JM, Eriksson N, Saxonov S, et al (2012). Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS ONE 7(4): e34267.
- Hill, WG & Weir, BS (2011). Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genetics Research, vol 93, no. 1, pp. 47-64. (See in particular Figure 5 which shows the distribution of actual genome sharing for different degrees of pedigree relationship.)
- Browning BL and Browning SR (2011). A fast, powerful method for detecting identity by descent. American Journal of Human Genetics 88(2):173-82. This paper includes a study on the comparative IBD sharing between the Welsh, Scottish and English.
- Huff CD, Witherspoon DJ, Simonson TS (2011). Maximum-likelihood estimation of recent shared ancestry (ERSA). Genome Research 21(5): 768-774.
- Powell JE, Visscher PM, Goodard ME (2010). Reconciling the analysis of IBD and IBS in complex trait studies. Nature Reviews Genetics 11, 800-805 (November 2010).
- Gusev A, Lowe JK, Stoffel M et al (2009). Whole population, genome-wide mapping of hidden relatedness. Genome Research 2009 19(2): 318-326. The GERMLINE algorithm for IBD detection.
- Visscher PM, Medland SE, Ferreira R et al (2006). Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLOS Genetics 2006 Mar; 2(3): e41. Epub 2006 Mar 24.
- Donnelly K (1983). The probability that related individuals share some section of genome identical by descent. Theoretical Population Biology 23(1): 34-63.
References
- ↑ Browning SR, Browning BL (2012). Identity by descent between distant relatives: detection and applications. Annual Review of Genetics 2012; 46: 617-33.
- ↑ Ralph P, Coop G (2013). The Geography of Recent Genetic Ancestry across Europe. PLOS Biology 11(5):e1001555.
- ↑ Data supplied by Tim Janzen. See Tim Janzen's presentation "Discovering and Verifying your Ancestry using Family Finder" at the 2014 Family Tree DNA Conference on Genetic Genealogy held in Houston, Texas, on 11 October 2014. The slides can be downloaded from http://tinyurl.com/p22ejo4
- ↑ Speed D, Balding DJ (2014). Relatedness in the post-genomic era. Nature Reviews Genetics 16: 33-34.
- ↑ For a detailed explanation see: Turner A. "Satiable Curiosity: Identity Crisis: Identical by State or Identical by Descent?" Journal of Genetic Genealogy Fall 2011, Volume 7.
- ↑ For an explanation of compound segments see the FTDNA FAQ I have tested my husband, our child, and myself. Why does our child have matches that neither parent has?
- ↑ Estes R. Parent-child non-matching autosomal DNA segments. DNAeXplained blog, 14 May 2015.
- ↑ Waterbury B. Father / mother/ child trio data. Rootsweb autosomal DNA mailing list, 6 October 2014.
- ↑ Kennett D. Father / mother/ child trio data. Rootsweb autosomal DNA mailing list, 7 October 2014.
- ↑ Ball CA et al (2020). "Matching White Paper: Discovering genetic matches across a massive, expanding genetic database".
- ↑ Durand EY, Eriksson N, McLean CY. Reducing pervasive false positive identical-by-descent segments detected by large-scale pedigree analysis. Molecular Biology and Evolution advance access publication online 30 April 2014.
- ↑ Pseudo/False Segments under 5 cM. Family Tree DNA Forum, 3 October 2015.
- ↑ 13.0 13.1 The information in this table was included in Tim Janzen's presentation "Discovering and Verifying your Ancestry using Family Finder" at the 2014 Family Tree DNA Conference on Genetic Genealogy held in Houston, Texas, on 11 October 2014. The slides can be downloaded from http://bit.ly/2EyT36N.
- ↑ See also the files on John Walden's website (now deprecated but archived at the Internet Wayback Machine) and particularly the chart atDNA segments that survive after phasing both sides.
- ↑ 15.0 15.1 Albrechtson A, Moltke I, Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics 2010; 186 (1 ):295-308. See Table 2 for the genomic positions of the regions on the different chromosomes where the peaks of IBD sharing were found.
- ↑ 16.0 16.1 Gusev A, Palamara PF, Apontel G et al. The architecture of long-range haplotypes shared within and across populations. Molecular Biology and Evolution (2011). doi: 10.1093/molbev/msr133. First published online: 6 October 2011. See Table 2 for details of the regions of high IBD sharing Annotated regions enriched for IBD sharing between HapMap populations.
- ↑ Gauvin H, Moreau C, Lefebvre J-F et al (2014). Genome-wide patterns of identity-by-descent sharing in the French Canadian founder population. European Journal of Human Genetics 22, 814–821.
- ↑ Turner A (2010). Up hill and down dale in the genomic landscape: the odd distribution of matching segments. Journal of Genetic Genealogy 6(1).
- ↑ 19.0 19.1 Li H, Glusman G, Hu H et al 2014. Relationship estimation from whole-genome sequence data. PLoS Genetics Jan 2014; 10(1): e1004144.
- ↑ Carmi S, Hui KY, Kochav E et al (2014). Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nature Communications 2014; 5: 4835. See in particular section 4.2 in the supplementary data pdf file where the authors discuss the IBD analysis.
- ↑ Chahine K. AncestryDNA matching update impacts Jewish ancestry. Ancestry blog, 1 August 2014.
- ↑ Swayne A. DNA matching just got better. Ancestry blog, 19 November 2014.
- ↑ Bettinger B. Goodbye false positives! AncestryDNA updates its matching algorithm. The Genetic Genealogist blog, 19 November 2014.
- ↑ Al-Khudhair A, Qiu S, Wyse M et al. Inference of distant genetic relations in humans using “1000 Genomes”. Genome Biology and Evolution (2015) doi: 10.1093/gbe/evv003. First published online: January 7, 2015.

